Lipopolysaccharide
Teichoic acids only occur in Gram-positive bacteria and lipopolysaccharide (LPS) only occur in Gram-negative bacteria. LPS is synthesized at the cytoplasmic and periplasmic leaflets of the inner membrane, and transferred to the outer membrane where it builds up a dense coat of polysaccharide chains at the surface of bacteria. Although the basic architecture of LPS is shared by most Gram-negative bacteria, a formidable structural heterogeneity is encountered across and even within bacterial strains under different growth conditions. LPS is essential for the growth of most Gram-negative bacteria by providing a barrier against environmental stress. In addition, the dense carbohydrate extensions of LPS protect bacteria from antibiotics and the immune system by blocking complement activation and antibody binding. The constant variation of terminal carbohydrate sequences, called O-antigens, also contribute to the evasion of bacteria from immune surveillance.
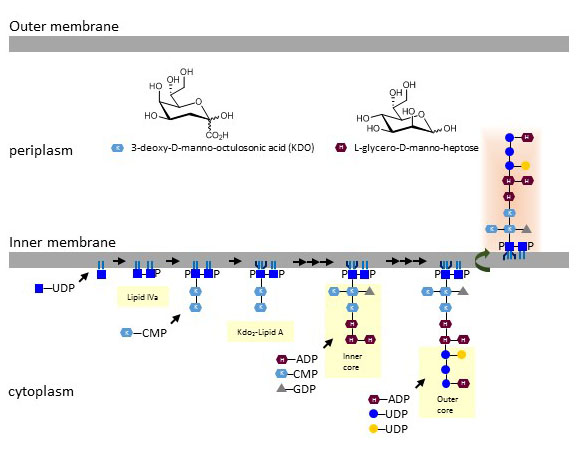
Structurally, LPS is subdivided in three components: lipid A, core oligosaccharide, and O-polysaccharide (O-antigen). Lipid A shows the highest structural conservation of the three LPS components across Gram-negative bacteria, although the number and type of acyl chains varies as well as the occurrence of multiple side-groups. To simplify the discussion, a generic LPS structure of Escherichia coli will be outlined here as example. LPS biosynthesis begins at the cytoplasmic side of the inner membrane by attaching two acyl chains to the C3 and to the de-acetylated amino group at C2 of UDP-GlcNAc. Two of such diacylated UDP-GlcNAc are combined through a β1-6 linkage and phosphorylated, thereby yielding the intermediate lipid IVa. Synthesis proceeds by addition of two units of the carbohydrate 3-deoxy-D-manno-octulosonic acid (abbreviated KDO). Lipid A is completed by the attachment of two or three additional acyl chains. Lipid A including the KDO disaccharide represent the minimal LPS backbone enabling the survival of Escherichia coli under laboratory conditions. Lipid A can be further modified by addition of phosphoethanolamine and 4-amino-4-deoxy-arabinose (L-Ara4N) side chains, which decrease the susceptibility of Escherichia coli to antibiotics of type polymyxin-B. The donor substrate for the transfer of L-Ara4N is not a nucleotide-activated sugar but undecaprenol-P-L-Ara4N. This donor substrate is analogous to dolichol-P-Man used in various eukaryotic glycosylation pathways.
The glycosylation of lipid A continues at the cytoplasmic leaflet of the inner membrane through the action of multiple glycosyltransferases using nucleotide-activated sugars as substrates. In Escherichia coli, the inner core includes L-glycero-D-mannoheptose (Hep), Gal, GlcN, GlcNAc, and Rha. The next group of oligosaccharides is heterogeneous and build at least five different “outer core” structures in Escherichia coli. The LPS core is then flipped to the periplasmic leaflet of the inner membrane, where subsequent elongations take place. Some Gram-negative bacteria further glycosylate LPS by addition of large O-polysaccharides, whereas other bacteria, such as Neisseria meningitidis and Campylobacter jejuni, have longer core oligosaccharides often including Sia but without O-polysaccharide addition. The resulting structures are then called lipooligosaccharides (LOS) to differentiate them from true LPS. Bacteria lacking outer core oligosaccharides show membrane instability and decreased pili expression. Such cells induce colanic acid expression, which is then cross-linked to LOS and favors the formation of biofilms
Figure X. Biosynthesis of lipid A and LPS core oligosaccharides (of Escherichia coli K12) at the inner cell membrane of Gram-negative bacteria.
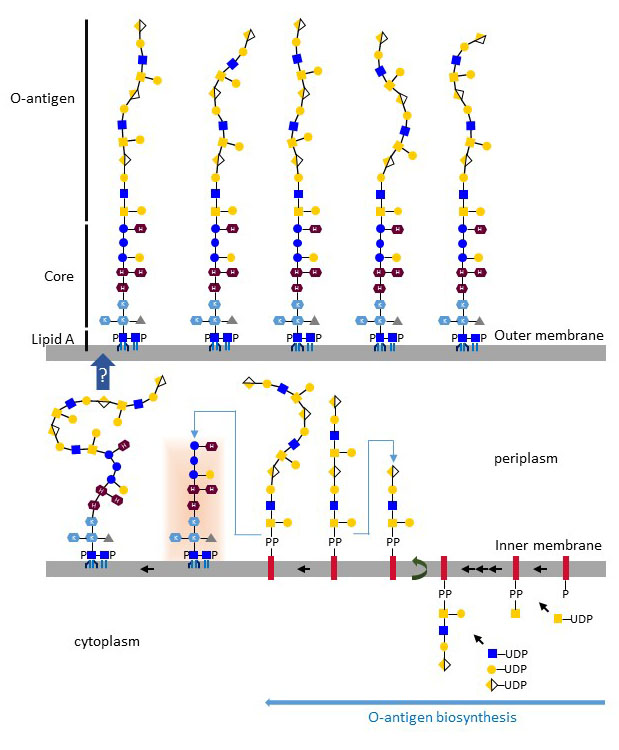
The biosynthesis O-polysaccharide units takes place on undecaprenol-PP at the cytoplasmic side of the inner membrane. About three to five monosaccharides are added by glycosyltransferases using nucleotide-activated sugar substrates. At this stage, the undecaprenol-PP-oligosaccharide is translocated to the periplasmic side of the inner membrane, where oligosaccharide units are ligated to a growing polysaccharide of repeating units. The typical complex controlling translocation, ligation and chain elongation are proteins of the WecA-Wzx-Wzy-Wzz family. Upon termination of chain extension, the O-polysaccharide is cleaved from undecaprenol-PP and transferred to the LPS core. The carbohydrate sequences of O-polysaccharides vary extensively; at least 60 different monosaccharides and 30 non-carbohydrate modifications have been identified to date. The structural complexity of O-polysaccharides is further increased by the multiple glycosidic linkages connecting monosaccharides. O-polysaccharides define the antigenic properties of Gram-negative bacteria. In Escherichia coli, about 170 O-serotypes have been described. After ligation of O-polysaccharide chains, LPS is transported by Lpt factors (LptBCFG, LptA, LptDE) through peptidoglycan and across the outer membrane, in which it decorates the external leaflet.
LPS is of tremendous medical importance as it is the main component of endotoxin. Even low doses of LPS in the range of 10 pg/ml activate leukocytes and induce a strong inflammatory response. LPS binds to the CD14/TLR4 complex, which is mainly expressed on leukocytes of the myeloid lineage. The endotoxic potency of LPS and its derivatives largely depend on the exposed organisms. For example, the phosphate groups on lipid IVa potentiate endotoxin activity on mouse leukocytes but prevent TLR4 activation on human leukocytes. The acyl chains of lipid A also influence the intensity of TLR4 activation. The LPS from the photosynthetic bacteria Rhodobacter sphaeroides even acts an endotoxin antagonist by competing for binding to the MD-2 protein, which is required for TLR4 activation by LPS. Rhodobacter sphaeroides LPS has only five acyl chains, which appears to be the key factor preventing TLR4 activation.
Figure X. Biosynthesis of LPS O-antigen (Escherichia coli O113) on undecaprenol-PP and transfer to the LPS core. Complete LPS molecules are transported to the outer membrane by the Lpt multiprotein complex.
The antibiotic polymyxin B kills Gram-negative bacteria by binding to LPS and destabilizing the outer membrane. The affinity of polymyxin B for LPS is applied in laboratory conditions to decontaminate reagents from LPS. The biosynthesis of LPS itself is not the target of known antibiotics, but several drug candidates have been shown to interfere with LPS biosynthetic enzymes, such as the GlcNAc deacetylase LpxC. The antibiotics tunicamycin and bacitracin inhibit the biosynthesis of O-polysaccharide through their interference with undecaprenol-PP formation and recycling.